[ad_1]
Modern investigate sheds light on the way an enzyme, included in getting old and metabolic regulation, interacts with our genetic information to control gene expression in just cells.
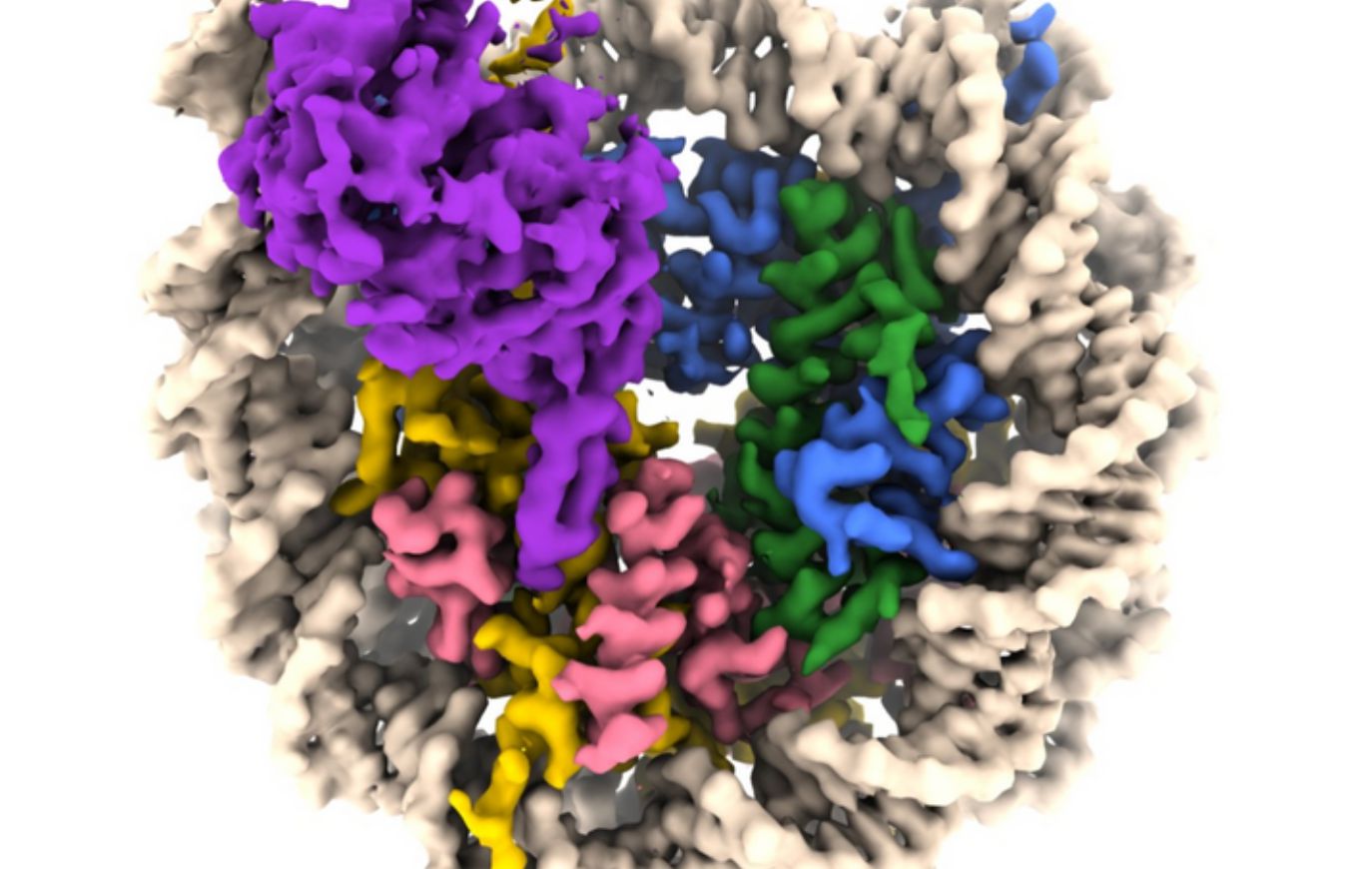
A team of researchers, spearheaded by a Penn Point out group, have captured visuals of a sirtuin enzyme connected to a nucleosome—a densely packed composition composed of DNA and histone proteins—demonstrating the enzyme’s capability to maneuver the nucleosome assembly and accessibility each DNA and histone proteins, thus elucidating its position in human beings and other organisms.
The conclusions are in depth in a paper revealed now in the journal Science Advances.
Sirtuins represent a course of enzymes existing in daily life varieties from germs to humans, with crucial functions in growing old, DNA destruction detection, and tumor suppression in several cancers. Owing to these numerous roles, the pharmaceutical marketplace is investigating their potential in biomedical applications. A considerable aim has been on the ability of specific sirtuins to lessen gene expression by getting rid of a chemical marker from histone proteins.
“In our cells,” as discussed by author Tune Tan, “DNA is not bare like we see it in textbooks it is spooled all-around proteins referred to as histones in a massive sophisticated named the nucleosome.”
The arrangement of this packaging can also influence gene activation or suppression: introducing an ‘acetyl’ chemical marker to the histone packaging product activates a gene, when getting rid of the acetyl marker deactivates it. Sirtuins can mute gene exercise by using absent the acetyl marker from histones inside of nucleosomes.
“Understanding how sirtuins interact with the nucleosome to take out this flag could inform long run drug discovery attempts.”
Prior research has generally examined how sirtuins engage with compact sections of histones separately, mainly because this kind of histone “tail” peptides are noticeably more simple to deal with in a laboratory location. Tan explains that nucleosomes are a hundred situations more substantial than the common histone peptides employed in these investigations, creating them significantly far more complex to perform with.
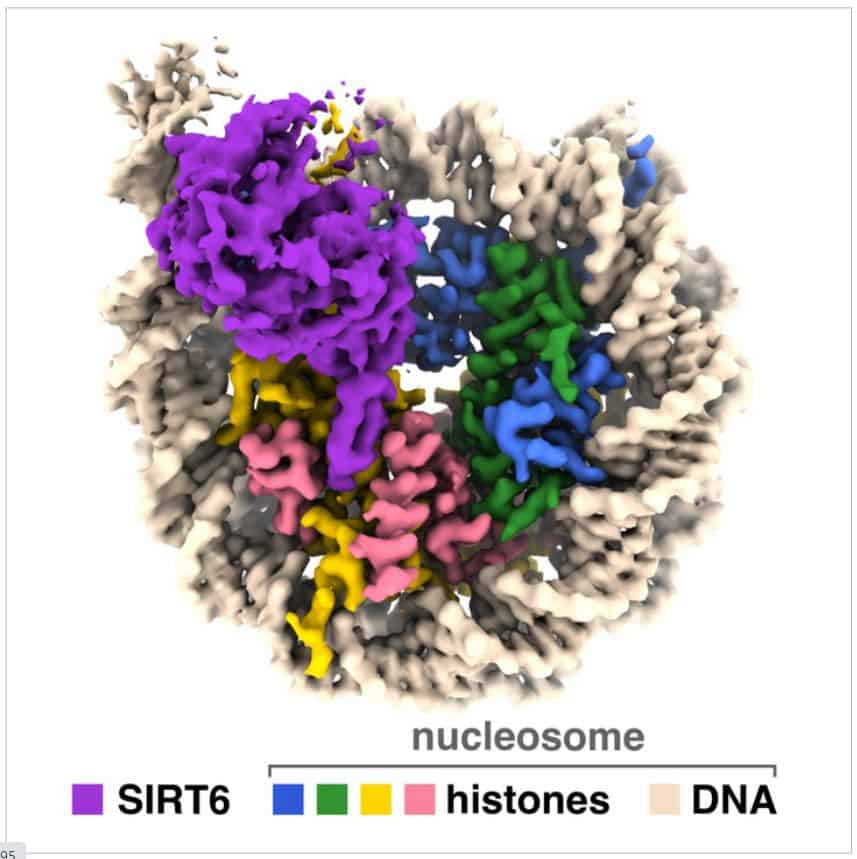
“We have visualized a sirtuin enzyme referred to as SIRT6 on its physiologically applicable substrate—the whole nucleosome,” adds creator Jean-Paul Armache.
They uncovered that “SIRT6 interacts with a number of sections of the nucleosome, not only the histone in which the acetyl flag is to be modified.”
Making use of a advanced imaging strategy named cryo-electron microscopy, with equipment at the Penn State Cryo-Electron Microscopy Facility, the Nationwide Cancer Institute, and the Pacific Northwest Cryo-EM Middle, the scientists pinpointed the way SIRT6 positions by itself on the nucleosome to clear away an acetyl team from the K9 area on the H3 histone. The staff collaborated with Craig Peterson’s lab at the College of Massachusetts Chan Professional medical Faculty to carry out biochemical experiments that corroborated their conclusions.
The researchers found out that SIRT6 attaches to the nucleosome through a link regarded as an “arginine anchor.” This sort of binding, 1st described by Tan’s lab in 2014, is employed by many proteins that target a notably acidic area on the nucleosome’s surface area. In this occasion, an prolonged loop—a structural feature of SIRT6—fits snugly into a melancholy in the acidic patch, significantly like a pipe resting in a trench.
“The arginine anchor is a typical paradigm for how numerous chromatin proteins interact with the nucleosome,” provides Tan. “When we mutated the SIRT6 arginine anchor, the action at the K9 placement was severely affected, supporting a crucial part for the SIRT6’s arginine anchor. Incredibly, this mutation also impacted SIRT6’s enzymatic activity at a distinct situation, K56, located substantially additional absent.”
Alternatively than SIRT6 attaching to the nucleosome utilizing two unique solutions to attain the two individual histone destinations, it appears to be that SIRT6 may possibly bind in a fashion that enables accessibility to K9, and concurrently gives obtain to K56.
“SIRT6 binds to a partially unwrapped nucleosome, with DNA displaced from the conclude of the nucleosome” adds Armache.
This revelation unveils the K56 posture, suggesting that SIRT6 might be equipped to primarily prolong alone to obtain that precise spot.
In the upcoming, the researchers aim to validate this speculation. They also hope to look into how SIRT6 cooperates with other enzymes and get a further knowledge of its function in the DNA hurt reaction approach.
Source: 10.1126/sciadv.adf7586
Picture Credit rating: Music Tan Lab, Penn State
[ad_2]
Supply hyperlink